Population Graphs: the graph theoretic shape of genetic structure
Dyer, R.J. and Nason, J.D. (2004), Population Graphs: the graph theoretic shape of genetic structure. Molecular Ecology, 13: 1713-1727. https://doi.org/10.1111/j.1365-294X.2004.02177.x
Abstract
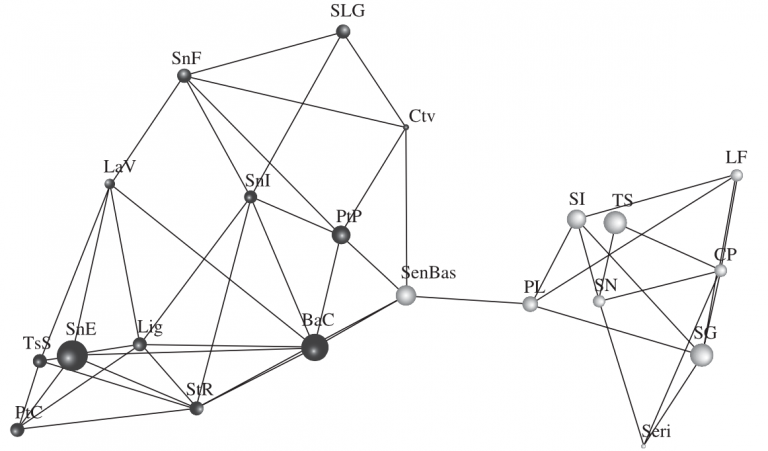
Patterns of intraspecific genetic variation result from interactions among both historical and contemporary evolutionary processes. Traditionally, population geneticists have used methods such as F-statistics, pairwise isolation by distance models, spatial autocorrelation, and coalescent models to analyze this variation and to gain insight into causal evolutionary processes. Here, we introduce a novel approach (Population Graphs) that analyzes marker-based population genetic data within a graph theoretical framework. This method can be used to estimate traditional population genetic summary statistics. Still, its primary focus is on characterizing the complex topology resulting from historical and contemporary genetic interactions among populations. We introduce the application of Population Graphs by examining the range-wide population genetic structure of a Sonoran Desert cactus (Lophocereus schottii). With this data set, we evaluate hypotheses regarding historical vicariance, isolation by distance, population-level assignment, and specific populations' importance to species-wide genetic connectivity. We close by discussing the applicability of Population Graphs for addressing a wide range of population genetic and phylogeographical problems.
